Waving nano-robots and molecular gearboxes that assemble themselves from a solution. That is what Professor Hendrik Dietz can already build with his nano-LEGO. So what’s next?
Professor Hendrik Dietz heads the laboratory for biomolecular nanotechnology at the Technical University München. His article in Science this spring opened up a box of construction elements on a molecular scale, all made from DNA.
Earlier this month (November 6th, 2015) Dietz visited the Kavli Institute of Nanoscience at the Faculty of Applied Sciences, where he reserved some time for an interview.
What inspired you to create this nano-construction kit with DNA?
“Just look at how all the macromolecular machinery is made in a cell, with shape-complementary components interacting with each other by weak contact interactions. If you want to make molecular machines synthetically, it seems you need to implement the same principles.”
Why did you choose DNA as the basis material?
“That’s our research object. We try to make molecular machines with DNA as construction material. We rely on DNA-based pairing as molecular glue to internally stabilise the objects. It makes the objects stable as a brick without interesting dynamics. We then plug multiple components together into a molecular machine with a well-defined structure and components that have pre-set degrees of freedom relative to each other.”
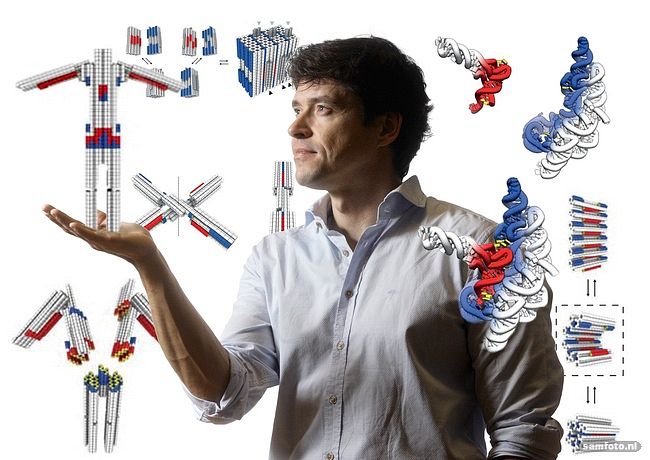
What kind of shapes and objects can you make in this way?
“We have made some little robot sculptures with moveable arms, and we built the smallest gear ever made. Now we’re working on integrating objects into more complex mechanisms, like a gearbox for example. In my talk, I showed a rotary mechanism with a ring, holding a rotor. It’s all 3D and self-assembling. I think no one has ever made something like that.”
How do you make these structures self-assembling?
“We exploit sequence complementarity between single strand DNA-molecules, like the string ATCG clings to TAGC. So when these single strands happen to bump into each other in solution, they start to form a double helix structure. If you have many of those partially complementary sequences, they link into branch-like geometries resulting in complex double-linked structures.
From there you can start making objects. I’m just basically sitting behind my computer to come up with nucleotide sequences for these molecules. Then we make the molecules, mix them and create a gear for example.”
How much time does the assembly take?
“If all the strands are in solution, you just stir it gently for about an hour, and it should be all right. It all depends on the complexity of the structure, but the assembly time is in that order for several hundreds of different DNA-strands with which we’re currently working. Macroscopically, an assembly through diffusion is hard to imagine. But it works nonetheless. As in any chemical process, the amount of material that is made depends on the raw material you put in at the start of the process. So we make a big tank with many of these molecules to make many of these gears all in parallel. It’s a scalable process to make nanostructures.”
It’s an amazing process. What kind of applications do you imagine?
“In the long run, we hope to make therapeutically relevant particles like drug-delivery vehicles that can intelligently identify target tissue or bacteria and release a drug on the spot. They may even open a cell pore to get the drug inside the cell. That may sound like science fiction, but these processes happen in nature all the time. Like viruses, for example, they do exactly that.”
So you could make artificial viruses?
“That’s the idea: to make viruses that are completely engineered and are all under control. Currently, people take a wild virus for gene delivery and try to make it harmless. That is complicated, and you don’t have all that much control over the natural virus. You can tinker a bit with the proteins, but you cannot change it completely. Our hope is that our technology can build drug-delivery viruses from the bottom up.
The other thing we’re trying to make is artificial enzymes, although that’s still a faraway goal. Enzymes are nature’s workhorses: they make biochemical processes happen. They make complex molecules out of simple components, or they break up large molecules into pieces. Enzymes do that wonderfully, much better than our chemistry does. If we could recreate that capability in a synthetic context, we could also do organic chemistry much better. Especially so in the synthesis of natural products, the way that plants do that. If we could design the right enzymes, it would allow us to do chemistry in a much more targeted and efficient way. That is a long-term goal.”
What is the short-term goal?
“In the short-term we’d like to use our objects as a tool in basic science. We use this technology now to study the interaction between protein molecules on a single-molecule level. In basic research, our tools allow researchers to do experiments at the molecular level.”
Cees Dekker, who invited you to the Kavli Institute, is working on constructing an artificial cell. Does your technology fit in with that project somehow?
“You could say there is a newly emerging field of synthetic biology where people are trying to re-engineer cells or create new cells. We are also synthetic biologists, but we try to recreate molecules from nature to understand how they work. A cell is full of macromolecular machines at which you can marvel. But I would like to build something like that because the process of building is the process of discovery and understanding. We prefer to make macromolecular machines; the cell is too complex for me.”
–> Hendrik Dietz c.s., Dynamic DNA devices and assemblies formed by shape-complementary, non-base paring 3D components, Science, March 27, 2015.


Comments are closed.